Enhancing signal detection capabilities beyond regular literature search
Methods and tools for data mining and all its variants, namely text mining and web mining, are emerging at cosmic speeds. But their implementation in pharmacovigilance and pharmacoepidemiology is still on its early stages.
The aim of this post is to explore and apply some of the current methods and tools using PubMed as the primary source for text mining. For this exercise I have chosen to mine PubMed abstracts for drug-induced acute kidney injury.
Searching for abstracts in PubMed
For this purpose, I used the PubMed Advanced Search Builder, which generated this search string: “(drug induced) AND acute kidney injury”, as shown here:
If you want to go directly to the results from that search, you can use https://www.ncbi.nlm.nih.gov/pubmed?term=(drug%20induced)%20AND%20acute%20kidney%20injury
At the time of writing this post, there were 8916 results from that search. The next step was to download all the abstracts into a text file, as shown on this screenshot:
Mining Abstracts with pubmed.mineR
Obviously, nobody has the time to read all the almost nine thousand abstracts. And if we had the time to do it, we would not have the ability, as human beings, to digest and integrate all this knowledge.
To help us with the task of knowledge discovery, we are going to use some applications in R language for the purpose of mining the text we have extracted. And this is when fun begins.
The R package we will use here is pubmed.mineR. The latest information on this package can be found here. To run the code I have used RStudio.
Package pubmed.mineR has many capabilities, most of them are not shown here. I have identified which of them would be more interesting for pharmacovigilance mining.
The initial code is shown below. In this post, code has a gray background, and the output a light blue background.
It starts by installing the package, and setting up the directory on your computer for input-output. I have used mine, but you will have to change it for your own path. The next step is to call the library.
# Install package:
install.packages(“pubmed.mineR”)
# Set directory:
setwd(“D:/PharmacovigilanceAnalytics.com/pubmed.mineR”)
# Call library(ies)
library(pubmed.mineR)
library(data.table)
# readabs will automatically read the abstracts from the pubmed file (pubmed_result.txt) and will write an S4 object which I named ‘akidrug’
akidrug <- readabs(“pubmed_result.txt”)
# printing first and last abstracts from akidrug:
printabs(akidrug)
The output resulting from ‘printabs(akidrug)’ is here, showing the first and the last abstracts:
Number of Abstracts 8916
Starts with
Renal Damaging Effect Elicited by Bicalutamide Therapy Uncovered Multiple Action Mechanisms As Evidenced by the Cell Model. Peng CC(1), Chen CY(2), Chen CR(3), Chen CJ(2), Shen KH(4)(5), Chen KC(6)(7)(8), Peng RY(9). Author information: (1)Graduate Institute of Clinical Medicine, School of Medicine, College of Medicine, Taipei Medical University, 250 Wu-Hsing Street, Taipei, 11031, Taiwan. (2)Wayland Academy, 101 North University Avenue, Beaver Dam, WI, 53916, USA. (3)International Medical Doctor Program, The Vita-Salute San Raffaele University, Via Olgettina 58, 20132, Milano, Italy. (4)Division of Urology, Department of Surgery, Chi Mei Medical Center, Tainan, 710, Taiwan. (5)Department of Optometry, College of Medicine and Life Science, Chung Hwa University of Medical Technology, Tainan, 717, Taiwan. (6)Graduate Institute of Clinical Medicine, School of Medicine, College of Medicine, Taipei Medical University, 250 Wu-Hsing Street, Taipei, 11031, Taiwan. kuanchou@tmu.edu.tw. (…
ADT-induced hypogonadism was reported to have the potential to lead to acute kidney injury (AKI).
ADT was also shown to induce bladder fibrosis via induction of the transforming growth factor (TGF)-β level.
Ends with
[APROPOS OF 8 CASES OF CARBON TETRACHLORIDE POISONING]. [Article in French] VEREERSTRAETEN P, VERNIORY A, VEREERSTRAETEN J, TOUSSAINT C, VERBANCK M, LAMBERT PP. NA NA
Word atomization
Something we can do is to determine the word frequency. For this purpose, pubmed.mineR uses “word_atomizations”:
akidrug_words <- word_atomizations(akidrug)
# Print the first 10 words by frequency
akidrug_words[1:10,]
The following table shows the first ten most frequent words. As expected, these most frequent words refer to the acute kidney injury aspect of your PubMed search. Please keep into account that word counting is one of the fundamental basis of text mining. Word counting contains still a very important research opportunity. I suggest to analyze, from the list generated by this example, word counts that are not as obvious as “renal”, “kidney”, or “patient” for this specific type of search.
| ID Number | Word | Frequency |
|---|---|---|
| 53805 | renal | 19824 |
| 18468 | acute | 9478 |
| 40387 | kidney | 8584 |
| 38691 | injury | 8236 |
| 49268 | patients | 7712 |
| 53138 | rats | 5519 |
| 32372 | failure | 5451 |
| 60217 | treatment | 4861 |
| 34861 | group | 4004 |
| 60509 | tubular | 3701 |
Gene atomization
Gene atomization will automatically fetch the genes (HGNC approved Symbol) from the text and report their frequencies.
# If you remember, akidrug is the name of the file for the collection of abstracts. Akidrug_gene will be the collection of genes found in those abstracts
akidrug_gene <- gene_atomization(akidrug)
# Next, we will obtain a subset of akidrug_gene containing 2 variables, one for the gene symbol and the other for the frequency
genes_table <- subset(akidrug_gene, select = c(“Gene_symbol”,”Freq”))
# Next, we prepare the whole gene database. The complete set can be obtained from the HGNC site.
hgnc<-read.delim(“D:/PharmacovigilanceAnalytics.com/pubmed.mineR/hgnc_complete_set.txt”,
header = T,stringsAsFactors = F)
We want to extract sentences containing Alias of the Human Genes, from the PubMed abstracts:
alias_fn(genes_table,hgnc,akidrug,”output”,c(“drug induced”,”acute kidney injury”,”adverse event”))
A sample from the results (saved to “outputalias”) is shown here:
TNF TNF-alpha
1
C3 C3b
PAH PH
PARP1 PARP
26184635
However, it is still unclear whether PARP overactivation happens during acute kidney injury (AKI) caused by endotoxic shock (ES).
And another one:
BAK1 BAK
2
CD5 T1
CR1 KN
ICAM1 CD54
IL18 IL-18
30531196
Other biomarkers of drug-induced kidney toxicity that have been detected in the urine of rodents or patients include IL-18 (interleukin-18), NGAL (neutrophil gelatinase-associated lipocalin), Netrin-1, liver type fatty acid binding protein (L-FABP), urinary exosomes, and TIMP2 (insulin-like growth factor -binding protein 7)/IGFBP7 (insulin-like growth factor binding protein 7), also known as NephroCheck®, the first FDA-approved biomarker testing platform to detect acute kidney injury (AKI) in patients.
- 1.Liu S, Liu J, Liu D, Wang X, Yang R. Inhibition of Poly-(ADP-Ribose) Polymerase Protects the Kidney in a Canine Model of Endotoxic Shock. Nephron. 2015;130(4):281-292. https://www.ncbi.nlm.nih.gov/pubmed/26184635.
- 2.Griffin B, Faubel S, Edelstein C. Biomarkers of drug-induced kidney toxicity. Ther Drug Monit. December 2018. https://www.ncbi.nlm.nih.gov/pubmed/30531196.
Literature Curation with PubTator Functionality
PubTator is a Web-based tool for accelerating manual literature curation (e.g. annotating biological entities and their relationships) through the use of advanced text-mining techniques. As an all-in-one system, PubTator provides one-stop service for annotating PubMed citations.
PubMed.mineR has a PubTator function. The PubTator function uses a PMID as entry and delivers results regarding chemicals, diseases, genes, and mutations, if they are referenced in the article. We are going to use the article by Griffin (see article 2 above, PIMD: 30531196) Let’s try it and see what hppens:
# Run PubTator function on PIMD 30531196 and save results on pubtator_output:
pubtator_output <- pubtator_function(30531196)
# Print PubTator output for chemicals, diseases, genes, and mutations:
pubtator_output$Chemicals
pubtator_output$Diseases
pubtator_output$Genes
pubtator_output$Mutations
Results are here:

There are many other pubmed.mineR functionalities. I encourage the reader to explore them and comment on the comments section of this post.
Exploration of other R packages.
Articles Published by Year and Word Cloud
This section is inspired on the code presented here.
library(RISmed)
library(dplyr)
library(ggplot2)
library(tidytext)
library(wordcloud)
result <- EUtilsSummary(“(drug induced) AND acute kidney injury”,
type = “esearch”,
db = “pubmed”,
datetype = “pdat”,
retmax = 30000,
mindate = 1960,
maxdate = 2019)
fetch <- EUtilsGet(result, type = “efetch”, db = “pubmed”)
abstracts <- data.frame(title = fetch@ArticleTitle,
abstract = fetch@AbstractText,
journal = fetch@Title,
DOI = fetch@PMID,
year = fetch@YearPubmed)
abstracts <- abstracts %>% mutate(abstract = as.character(abstract))
abstracts %>%
head()
abstracts %>%
group_by(year) %>%
count() %>%
filter(year > 1959) %>%
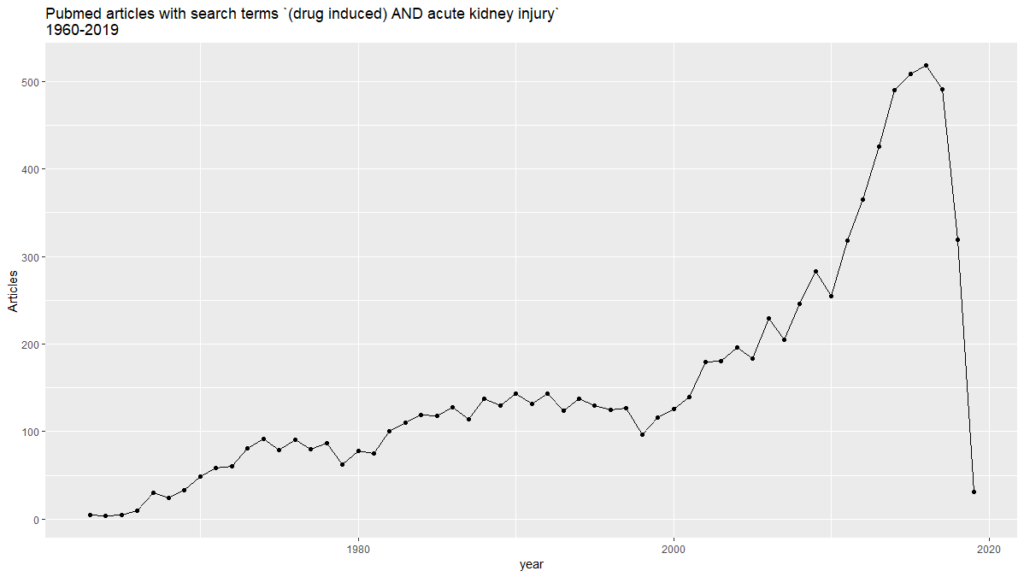
ggplot(aes(year, n)) +
geom_point() +
geom_line() +
labs(title = “Pubmed articles with search terms (drug induced) AND acute kidney injury \n1960-2019″, hjust = 0.5,
y = “Articles”)
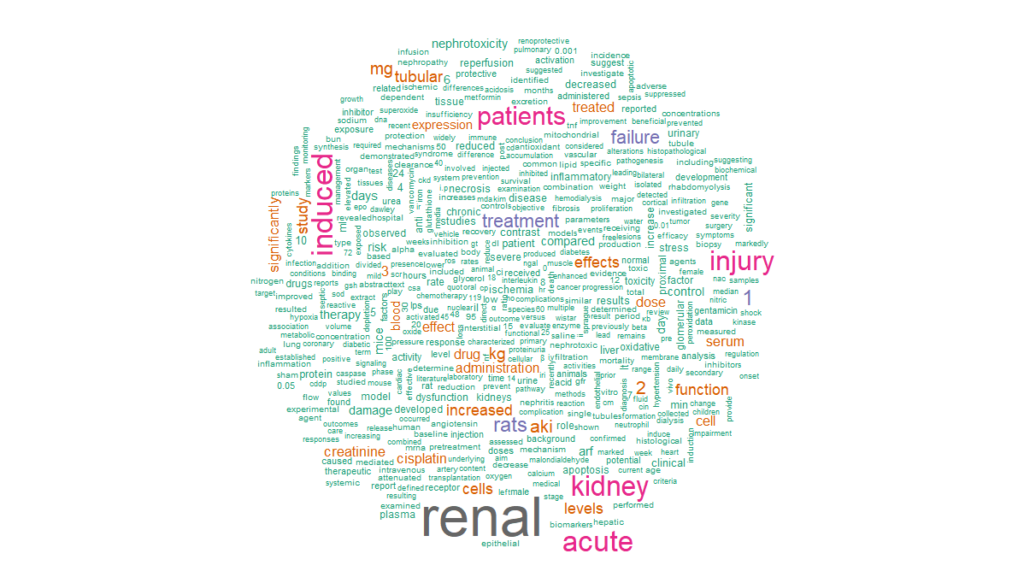
cloud <- abstracts %>%
unnest_tokens(word, abstract) %>%
anti_join(stop_words) %>%
count(word, sort = TRUE)
cloud %>%
with(wordcloud(word, n, min.freq = 15, max.words = 500, colors = brewer.pal(8, “Dark2”)), scale = c(8,.3), per.rot = 0.4)
This is the first of a series of posts analyzing text mining applications for PubMed. The second one explores knowledge graphs and semantic analytics.